A deep dive into the proteins and protein activities of human tumors grown in mouse models expands the potential therapeutic pathways to treat medulloblastoma, according to a new study published in the Journal of Proteome Research.
As part of the study, the collaboration between researchers at the Translational Genomics Research Institute (TGen), part of City of Hope, Sanford Burnham Prebys Medical Discovery Institute and the University of California, San Diego provided a mechanistic explanation for why the drug actinomycin D might be a useful candidate for treating Group 3 medulloblastoma—building on findings from a previous drug screening study.
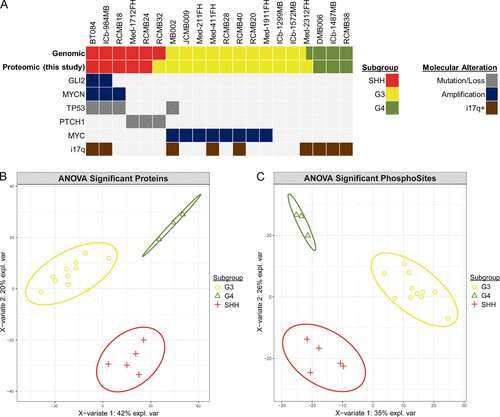
Medulloblastoma, the most common malignant pediatric brain tumor, has four main molecular subtypes. Group 3 tumors have a poor prognosis, with five-year survival rates of 50%. There is a significant need for more effective treatments and for treatments with less toxic side effects for all medulloblastoma, the researchers note.
To look for new treatment targets, TGen Postdoctoral Fellow Kristin Leskoske, Ph.D. and her colleagues studied tumors that were grown in mice from cells taken from 20 medulloblastoma patients with different subtypes of the disease. The resulting tumors are called orthotopic patient-derived xenografts or PDX models.
“There has been some debate in the field about whether PDX models of medulloblastoma are close enough to primary human tumors for research and drug testing,” said Patrick Pirrotte, Ph.D., TGen Associate Professor, Director of the Collaborative Center for Translational Mass Spectrometry, and the paper’s senior author.
One of the important conclusions of the study, he added, is “that from a proteomics perspective, these PDX tumors are very similar to actual patient tumors and can be used as models of the disease.”
Cells grown directly in the mouse brain, rather than in a dish in the lab or as an implant in the mouse flank as is sometimes done, “more closely mimic the environment that a tumor normally grows in,” said Dr. Leskoske, the paper’s first author.
The researchers then deeply profiled the medulloblastoma proteome, measuring the amount of each protein made in each PDX model and identifying chemical modifications on the proteins, called phosphorylations, that indicate the protein’s activity level.
Using these protein profiles, Dr. Leskoske and colleagues were able to correlate the abundance or activity of certain proteins with their sensitivity to particular drugs. For instance, a previous study by co-author Robert J. Wechsler-Reya, Ph.D., of Sanford Burnham Prebys Medical Discovery Institute suggested Group 3 medulloblastoma was sensitive to actinomycin D, but the reasons for this sensitivity were unclear.
“Drug screens identify vulnerabilities; they don’t identify their mechanism of action. Our complementary proteomic study proposes a potential mechanism for actinomycin D sensitivity,” said Dr. Pirrotte.
The most common molecular feature of Group 3 medulloblastoma is amplification of MYC. In this study, the researchers found a significant association between actinomycin D sensitivity and an increased abundance of MYC and genes targeted by MYC.
Many of the proteins in the study that were sensitive to actinomycin D are mitochondrial proteins, which are key in regulating cell metabolism. The team has other unpublished data that, combined with these findings, suggest that certain metabolic vulnerabilities could be targeted in Group 3 tumors, Dr. Pirrotte said.
Source: Read Full Article
