A reliable method to assess infection with the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in vaccinated individuals has yet to be established. The development of a reliable diagnostic tool for the identification of recent and remote SARS-CoV-2 infection is needed to enable population-based seroprevalence studies, and diagnose post-infectious complications of the coronavirus disease 2019 (COVID-19), and ascertain infection status in vaccine clinical trials.
Study: Anti-nucleocapsid antibodies following SARS-CoV-2 infection in the blinded phase of the mRNA1273 Covid-19 vaccine efficacy clinical trial. Image Credit: ustas7777777 / Shutterstock.com
Background
Current COVID-19 vaccines do not elicit anti-nucleocapsid (N) antibodies.
Comparatively, many studies have successfully detected the presence of anti-N antibodies in unvaccinated individuals with high sensitivity and specificity approximately 14 days post-infection. In fact, the anti-N antibody response in unvaccinated individuals appears to be quite durable, with a half-life ranging from 68 to 283 days.
Previous SARS-CoV-2 infections in participants in pivotal Phase III clinical trials and various seroprevalence studies that were carried out during the early stages of the pandemic relied on the detection of anti-spike or anti-N antibodies through chemiluminescence immunoassays in unvaccinated individuals. However, the extent to which vaccine-induced immunity impacts seroconversion to non-spike proteins and whether these types of immunoassays can be used to determine previous SARS-CoV-2 infections in regions with high vaccine coverage is unknown.
A new study posted to the preprint server medRxiv* evaluates vaccine and placebo recipients in the mRNA-1273 Phase III clinical trial with a history of COVID-19 during the blinded placebo-controlled phase of the trial. The current study assayed anti-N antibodies from the serum samples of the participants for five months post-enrollment.
About the study
The current study enrolled 30,420 U.S. adults from the COVE trial who were 18 years of age or above between July 27, 2020, and October 23, 2020, and was continued until March 26, 2021.
All study participants received two 100 micrograms (µg) doses of either mRNA-1273 vaccine or placebo on Day 1 (baseline) and Day 29. Detection of previous SARS-CoV-2 infection was based on both symptom-prompted testing and planned diagnostic testing at specific study visits. Any participant who met predefined clinical criteria for COVID-19 during follow-ups underwent an illness visit for the collection of a nasopharyngeal (NP) swab for testing.
Blood samples were collected from the participants for anti-N antibody serology on Day 1, Day 29, Day 57, and at the Participant Decision Visit (PDV). PDV is when participants are informed about their randomization assignment and administered mRNA-1273 vaccination if they were previously assigned to placebo.
Detection of anti-N antibodies was achieved through the use of the Roche Elecsys Anti-SARS-CoV-2 immunoassay. NP swabs for SARS-CoV-2 reverse transcription-polymerase chain reaction (RT-PCR) testing were collected from all participants on Day 1, Day 29, and the PDV.
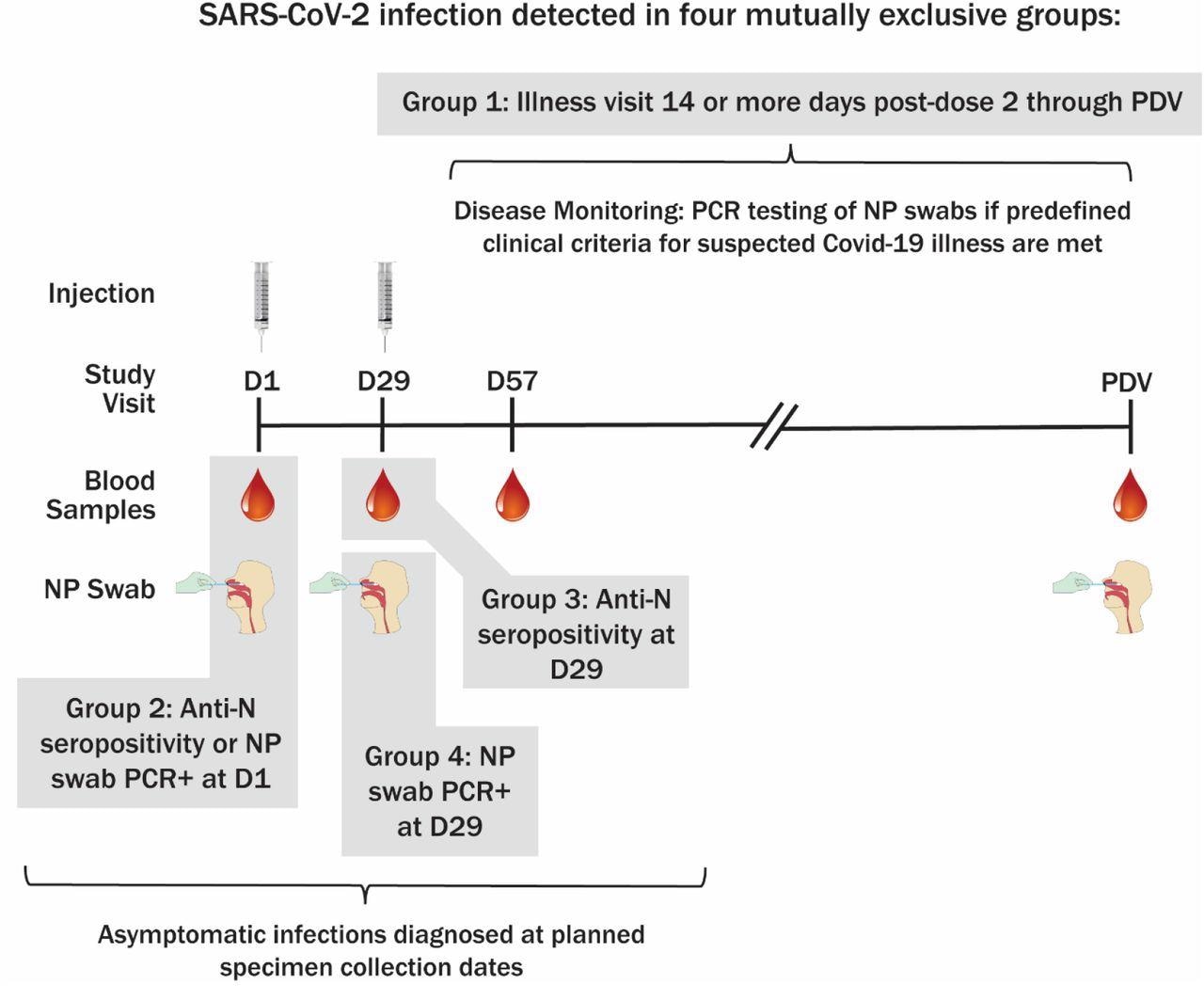
Method of SARS-CoV-2 infection determination and sampling schedule. PDV, participant decision visit.
Viral copy number was assessed for confirmed COVID-19 cases at illness visits Day 1, 3, 5, 7, 9, 14, 21, and 28 by RT–PCR as well as the conversion of cycle-threshold (Ct) values to viral genome copy number.
Four mutually exclusive groups of infection were defined by the study. The first group consisted of primary endpoint COVID-19 cases that were detected at illness visits 14 days post-second vaccine dose until the PDV, whereas the second group consisted of study participants who were anti-N seropositive or NP positive at baseline.
The third group consisted of cases that were anti-N seropositive at Day 29 without any evidence of prior infection, whereas the fourth group included cases that were NP positive but seronegative at Day 29 without any evidence of prior infection.
COVID-19 cases were defined as baseline SARS-CoV-2 negative in participants with at least two SARS-CoV-2 symptoms and one swab sample that was positive for SARS-CoV-2 by RT-PCR.
Study findings
SARS-CoV-2 infection was detected at baseline in 684 participants, anti-N seropositivity was detected on Day 29 in 107 participants, and NP positivity was detected on Day 29 in 49 participants. Additionally, 812 baseline SARS-CoV-2 negative participants were found to experience illness 14 or more days after receiving the full dose of the vaccine or placebo.
The anti-N seropositivity at the PDV was 40.4% for vaccine recipient COVID-19 cases and 93.3% for placebo recipient COVID-19 cases. The 50% inhibitory dilution (ID50) neutralizing antibody titers were similar in anti-N seronegative and anti-N seropositive at PDV for the vaccine recipients.
Furthermore, the median viral copy number was higher in placebo recipients at Day 1 illness visit who were anti-N seropositive on the PDV as compared to those who were anti-N seronegative. Anti-N seropositivity was 13.67 times higher for placebo as compared to the vaccine for any given viral copy number. However, similar anti-N seropositivity rates were observed on Day 29 for both the vaccine and placebo groups.
Day 29 and PDV anti-N seropositivity rates were lower for NP-positive participants as compared to baseline anti-N seropositive participants. Additionally, Day 57 and PDV anti-N seropositivity rates were lower for Day 29 NP positive and anti-N seronegative participants as compared to Day 29 anti-N seropositive participants.
Conclusions
The current study indicated that anti-N antibodies may have lower sensitivity in mRNA-1273-vaccinated individuals who were previously infected with SARS-CoV-2. Further research is needed to determine SARS-CoV-2 infections in a population through serosurveillance in the era of COVID-19 vaccination coverage.
Vaccination status must be considered while interpreting seroprevalence and seropositivity data that is solely based on anti-N antibody testing.
Limitations
The study had certain limitations. First, the participants received only the mRNA-1273 vaccine. Second, the study uses only Elecsys assay, which has the highest sensitivity and it is unknown whether other immunoassays will have different sensitivity.
A third, limitation was the small sample size of the study. Additionally, most SARS-CoV-2 infections following vaccinations are caused by Delta and Omicron variants, both of which are associated with a high viral copy number. Finally, the current study did not address any pre-specified objective of the COVE study.
*Important notice
medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Follmann, D., Janes, H. E., Buhule, O. D., et al. (2022). Anti-nucleocapsid antibodies following SARS-CoV-2 infection in the blinded phase of the mRNA1273 Covid-19 vaccine efficacy clinical trial. medRxiv. doi:10.1101/2022.04.18.22271936. https://www.medrxiv.org/content/10.1101/2022.04.18.22271936v1.
Posted in: Medical Science News | Medical Research News | Disease/Infection News
Tags: Antibodies, Antibody, Assay, Blood, Clinical Trial, Coronavirus, Coronavirus Disease COVID-19, CT, Diagnostic, Diagnostics, Efficacy, Genome, immunity, Immunoassay, Immunoassays, Nasopharyngeal, Omicron, Pandemic, Placebo, Polymerase, Polymerase Chain Reaction, Research, Respiratory, SARS, SARS-CoV-2, Serology, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Syndrome, Transcription, Vaccine

Written by
Suchandrima Bhowmik
Suchandrima has a Bachelor of Science (B.Sc.) degree in Microbiology and a Master of Science (M.Sc.) degree in Microbiology from the University of Calcutta, India. The study of health and diseases was always very important to her. In addition to Microbiology, she also gained extensive knowledge in Biochemistry, Immunology, Medical Microbiology, Metabolism, and Biotechnology as part of her master's degree.
Source: Read Full Article