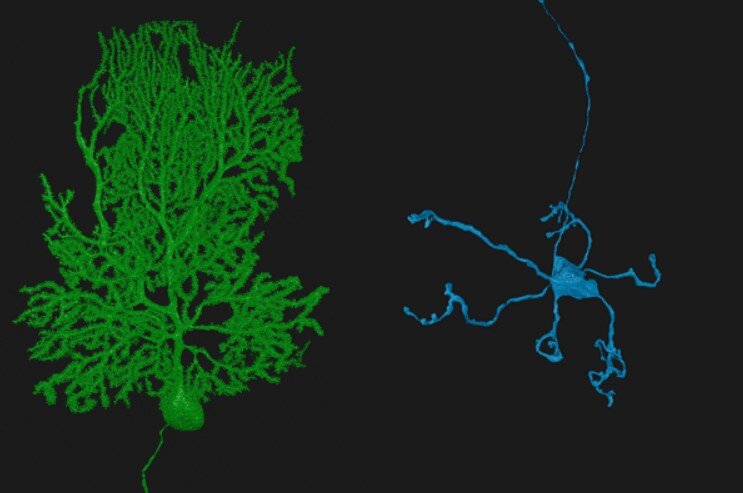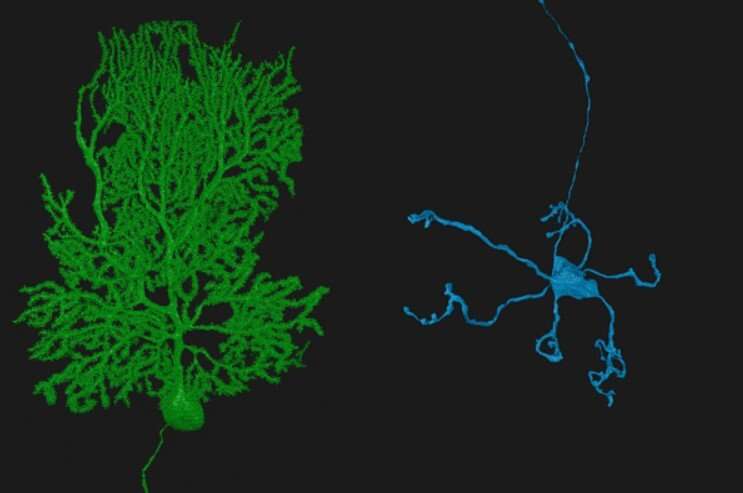
Many of us have seen microscopic images of neurons in the brain—each neuron appearing as a glowing cell in a vast sea of blackness. This image is misleading: Neurons don’t exist in isolation. In the human brain, some 86 billion neurons form 100 trillion connections to each other—numbers that, ironically, are far too large for the human brain to fathom.
Wei-Chung Allen Lee, Harvard Medical School associate professor of neurology at Boston Children’s Hospital, is working in a new field of neuroscience called connectomics, which aims to comprehensively map connections between neurons in the brain.
“The brain is structured so that each neuron is connected to thousands of other neurons, and so to understand what a single neuron is doing, ideally you study it within the context of the rest of the neural network,” Lee explained.
Lee recently spoke to Harvard Medicine News about the promise of connectomics. He also described his own research, which combines connectomics with information on neural activity to explore neural circuits that underlie behavior.
Harvard Medicine News: To start with a basic question, what is connectomics?
Lee: We define connectomics as understanding how individual neurons are connected to one another to form functional networks. The goal is to create connectomes, or detailed structural maps of connectivity where we can see every neuron and every connection. What’s unique is the comprehensiveness of connectivity: In a perfect connectome, we’d know how every neuron was connected to every other neuron.
We believe that the connectivity of neurons is fundamental to how they function, since they must receive information from each other in order to use this information. Having comprehensive data about connectivity allows us to look at higher-order interactions between populations of neurons that are important for brain function and behavior. It is challenging to study higher-order interactions without connectomics.
Some have argued that you are your connectome. When you fall asleep at night, your brain activity dramatically changes, interrupting your thoughts and feelings—but when you wake up, you resume your thoughts and feelings without any break in your sense of self. This is likely because your brain connectivity has remained largely intact through the night. In essence, the structure of how our neurons are wired is our “self,” and connectomics is the key to understanding this structure.
What are you studying within the context of connectomics?
My lab is interested in understanding how computations arise in the brain, or the general principles by which neural circuits organize themselves into functional networks. To do this, we aim to comprehensively map how individual neurons are connected to one another in complex networks. At the same time, we want to understand how those neurons are active within the functioning circuit. We do this in the context of behavior, ranging from making decisions to executing actions.
We are trying to couple connectomics with recordings of neural activity to do what we call functional connectomics. Essentially, we take the map of where every neuron is and how it is connected to every other neuron, and we layer on information about the activity of those neurons in a living animal. We also use genetic engineering approaches to label specific cell types, which is additional information that we can layer on top of connectivity.
What tools do scientists use to map connectomes?
We are developing and applying high-throughput microscopy, computational approaches, and machine learning to generate connectomes and translate these detailed maps of neural connectivity into biological and computational insights. One key component of our approach is serial transmission electron microscopy, or EM, which has unsurpassed spatial resolution, signal-to-noise ratio, and speed relative to other serial EM methods. This technique allows us to identify excitatory and inhibitory neurons, as well as the synapses, or small gaps where neurons connect to each other. We can also examine connectivity patterns of neurons, and study the organization of synaptic connections.
Historically, high-resolution EM has been slow and tedious, but we’ve engineered a high-speed EM platform that allows us to capture the whole nervous system of an adult fruit fly in a few months, generating 5 to 10 terabytes of data a day. We have also developed computational infrastructure and tools that enable us to handle and visualize the large amounts of data that we are generating. For example, we use artificial deep neural networks to extract information about cells and their connectivity from these massive datasets.
What models do you use in your research?
We have mainly worked with mice and fruit flies, which are powerful and well-studied model systems. The field has sophisticated genetic tools that allow us to label different populations of neurons across the central nervous systems of these species. In fruit flies, we can use the technologies we’ve been developing for connectomics to capture the entire brain and nervous system at synapse resolution. In the mouse, we can target relevant neural circuits or subcircuits. We are using these models to study the basic principles of how neural circuits are built and operate—basically how neural networks are connected to each other to perform different computations that underlie behavior.
We also work in nontraditional model systems such as the mosquito. Mosquito brains are about the same size as fruit fly brains, but the genetics is more challenging. Scientists have used genetics to access the first-order neurons that start carrying information into the mosquito brain, but the rest of the brain is a black box in many respects. We don’t know much about its fundamental neurobiology, including how the mosquito brain integrates different sensory modalities to drive behavior.
For example, adult female mosquitoes that are trying to reproduce integrate information on human odors, heat, and carbon dioxide. We know that these different sensory cues enter the brain, but we don’t know how they are integrated and converge onto neural circuits that drive mosquitoes’ host-seeking behavior.
We hope that mapping the whole mosquito brain will provide a new foundation for understanding how sensory integration and action selection works for innate behavior. Additionally, the specific mosquito species we study is a vector for diseases such as malaria, West Nile, Zika, yellow fever, and dengue fever, so there’s a clinical and public health aspect of this that makes it a really important model system.
You recently published a paper in Nature on brain connectivity and pattern association in mice. What was the premise of the study?
This was a collaboration with Wade Regehr, professor of neurobiology at HMS. The paper focuses on information processing in the cerebellum, which is a brain region that, among other things, is important for smooth, coordinated movement. One of the things the cerebellum is thought to do is make fine-scale error corrections in movement by comparing patterns from intended and executed actions. For example, if you try to touch your nose and you miss, there is information coming from your motor system that tells your cerebellum what the intended action was, and there is sensory information coming from your finger about what actually happened, including the location of your finger in space. The cerebellum is thought to compute the difference between the intended action and the actual action, and to help correct the error.
We studied the cerebellar cortex, which is packed full of small neurons called granule cells that make up more than half the neurons in the brain. These granule cells each have, on average, four dendrites, or branching structures that receive information from other neurons. In this case, the dendrites connect to neurons called mossy fibers that bring information into the cerebellum. The granule cells then process this information and communicate it to other neurons called Purkinje cells, with each Purkinje cell integrating information from 100,000 to 200,000 granule cells, and sending this information to other brain regions. These three cell types make up the “feedforward” circuit we wanted to better understand.
What was your key finding in the Nature paper?
Previously, scientists and computational models assumed that the dendrites on granule cells randomly connected to different mossy fibers, and this randomness contributed to the complexity and encoding capacity of the information communicated to Purkinje cells. However, using connectomics, we mapped the connections between mossy fibers, granule cells, and Purkinje cells. We found that the dendrites on granule cells don’t connect to mossy fibers in a random way. Instead, they connect to mossy fibers selectively, with more granule cells connecting to the same mossy fibers than expected. This selectivity should decrease the encoding capacity of the information that can be conveyed—but it turns out that for only a very small decrease in capacity, you get more robustness in pattern association. We think this is because there is more redundancy in the connections between granule cells and mossy fibers, and granule cells may be connecting to more-informative mossy fibers.
This is a finding that leverages connectomics to establish more comprehensive circuit structure by allowing us to look at how large populations of neurons are connected to each other in the same circuit. We need this connectivity information to make detailed and comprehensive models of how information flows through the network. This paper demonstrates how connectomics can be used to provide data to test long-standing theories about information processing and complex neural networks.
What else do you think connectomics can help scientists figure out?
Something that I think is going to be really powerful in the near future is what people are calling “comparative connectomics,” or comparing different connectomes. I’m particularly excited about looking at how behavioral differences across individuals correlate with differences in their connectomes. I’m also interested in comparing connectomes for different species to see what principles are conserved in different kinds of brains. In addition to finding conserved principles that can be generalized across species, I want to find differentiating principles that make humans unique. Ultimately, our common humanity may lie in the shared structure of how our brains are wired.
Why do you think connectomics is such a growing field?
Progress has been in part driven by advances in technology, including advances in mechanical engineering that allow us to scale data acquisition, as well as advances in genetic engineering that allow us to label specific cell types. Additionally, the field has been transformed by machine learning, which can be used to analyze these datasets to extract biological insight. The connectomics field is an interesting convergence of neurobiology, engineering, computing power, and artificial intelligence.
We’ve been developing a lot of different technologies for scaling up data generation and data analysis that I think will be useful in other scientific disciplines. We’re generating some of the biggest image datasets in the world right now, and there are more to come. For example, the NIH has a goal of mapping a whole mouse brain connectome in the next 10 years, which would be about a zettabyte of data, or a trillion gigabytes. Researchers also want to map human and nonhuman primate brains.
We’ve only scratched the surface of understanding how neurons are connected to one another to form functional networks, but connectomics is arguably transforming neuroscience. I believe we are on the cusp of understanding circuit mechanisms underlying how neurons and networks of neurons compute. We’re on the precipice of understanding the basic building blocks of neural networks, including the rules by which they connect to one another and the rules that underlie the computations they carry out. To me, that is really, really exciting.
More information:
Tri M. Nguyen et al, Structured cerebellar connectivity supports resilient pattern separation, Nature (2022). DOI: 10.1038/s41586-022-05471-w
Journal information:
Nature
Source: Read Full Article