Biology is rife with examples of collective behavior, from flocks of birds and colonies of bacteria to schools of fish and mobs of people. In a study with implications from oncology to ecology, researchers from Rice University and the University of Georgia have shown that data science can unlock subtle clues about the individual origins of collective behavior.
When a group of individuals move in synch, they can create patterns—like the flocking of birds or “the wave” in a sports stadium—that no single individual could make. While these emergent behaviors can be fascinating, it can be difficult for scientists to zero in on the individual actions that bring them about.
“You see emergent behaviors by looking at the group rather than the individual,” said Rice bioengineer Oleg Igoshin, a theoretical biophysicist who has spent almost 20 years studying emergent behavior in cells—be they cooperative bacteria, cancer cells or others.
In a study published online this week in the American Society for Microbiology journal mSystems, Igoshin and Rice alumnus Zhaoyang Zhang developed a method to assess which aspects of individual behavior give rise to emergent behavior.
To illustrate both the difficulty and the importance of understanding these individual contributions to the collective group, Igoshin uses the example of metastatic cancer, where a group of cells with a particular mutation are moving toward the surface of a tumor so they can break away and form a new tumor elsewhere.
“Most of these cells fail to escape the original tumor, and the question is, what determines which ones will succeed?” asked Igoshin, a professor of bioengineering and senior scientist at Rice’s Center for Theoretical Biological Physics. “What property is a signal for emergence? Is it how fast they move? Is it how long they move before changing direction? Perhaps it’s how frequently they stop. Or it could be a combination of several signals, each of which is too weak to bring about emergence on its own but which act to reinforce one another.”
As a group, the potentially metastatic cancer cells share some key traits and abilities, but as individuals, their performance can vary. And in a large population of cells, these performance differences can be as stark as those between Olympic athletes and couch potatoes. Above all, it is this natural variation in individual performance, or heterogeneity, that makes it so difficult to zero in on individual behaviors that contribute to emergent behaviors, Igoshin said.
“Even for cells in a genetically homogeneous tumor, if you look at individuals there will be a distribution, some heterogeneity in performance that arises from some individuals performing 50% above average and others 50% below average,” he said. “So the question is, ‘With all of this background noise, how can we find the weak trends or signals associated with emergence?'”

Igoshin said the new method incorporates data science to overcome some weaknesses of traditional modeling. By populating their models with experimental data about the movements of individual cells, Igoshin said he and Zhang, who received his Ph.D. from Rice in May, simplified the search for individual behaviors that influence group behaviors.
To demonstrate the technique, they partnered with Lawrence Shimkets, whose lab at the University of Georgia (UGA) has spent years compiling data about the individual and group behaviors of the cooperative soil bacterium Myxococcus xanthus.
“They’re predatory microbes, but they are smaller than many of the things they eat,” Igoshin said of M. xanthus. “They work together, kind of like a wolf pack, to surround their prey and make the chemicals that will kill it and digest it outside of their bodies, turning it into molecules that are small enough for them to take in.”
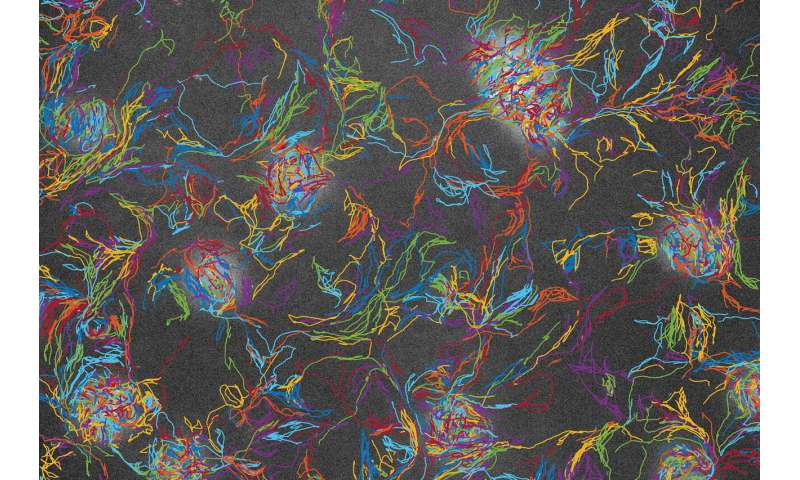
During times of stress, like when food is running short, M. xanthus exhibit a form of emergent behavior that has been studied for decades. Like lines of cars flowing into a city at rush hour, they stream together to form densely packed mounds that are large enough to see with the naked eye. Mound formation is an early step in the process of forming rugged long-lived spores that can reestablish the colony when conditions improve.
In a previous study, UGA’s Chris Cotter, a co-author of the new study and a graduate student in Shimkets’ group, tracked individual behaviors of wild-type cells and collaborated with Igoshin to develop a data-driven model that uncovers the cellular behaviors that are key to aggregation. In the new study, Cotter and Zhe Lyu, a former UGA postdoctoral researcher now at Baylor College of Medicine in Houston, collected mound-forming data from mixtures of three strains of M. xanthus: a naturally occurring wild type and two mutants. On their own, the mutants were incapable of forming mounds. But when a significant number of wild-type cells were mixed with the mutants, they were “rescued,” meaning they integrated with the collective and took part in mound-building.
“One of the mutants is fully rescued while the other is only partially rescued, and the goal is to understand how rescue works,” Igoshin said. “When we applied the methodology, we saw several things that were unexpected. For example, for the mutant that’s fully rescued, you might expect that it behaves normally, meaning that all its properties—its speeds, its behaviors—will be exactly the same as the wild type. But that’s not the case. What we found was that the mutant performed better than normal in some respects and worse in others. And those compensated for one another so that it appeared to be behaving normally.”
Emergent behaviors in M. xanthus are well-studied, classic examples. In addition to showing that their method can unlock some of the mysteries of M. xanthus’ behavior, Igoshin said the new study indicates the method can be used to investigate other emergent behaviors, including those implicated in diseases and birth defects.
Source: Read Full Article
