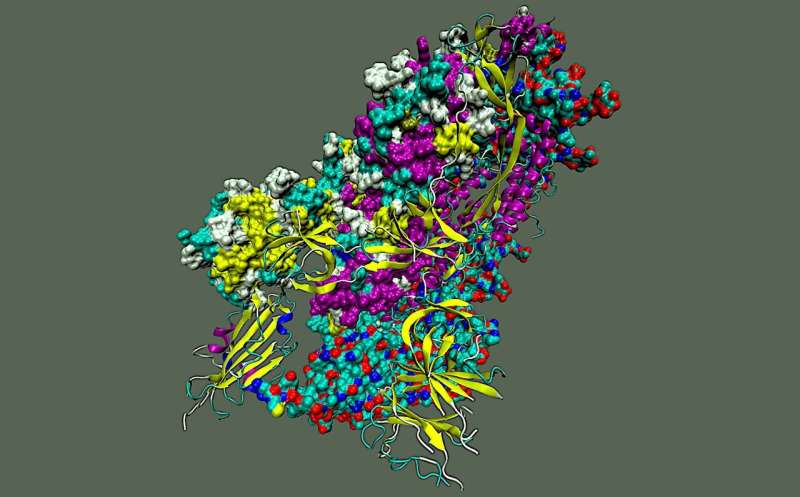
To explore the inner workings of severe acute respiratory syndrome coronavirus 2, or SARS-CoV-2, researchers from the Department of Energy’s Oak Ridge National Laboratory developed a novel technique.
The team—including computational scientists Debsindhu Bhowmik, Serena Chen and John Gounley—ran molecular dynamics simulations of the novel virus that caused the COVID-19 disease pandemic on ORNL’s Summit supercomputer, an IBM AC922 system. The researchers then analyzed the output with a customized deep learning approach to produce a complete molecular picture of the “spike” protein on the virus’s surface.
This method enabled them to pinpoint specific flexible regions, which they studied in extreme detail to reveal promising therapeutic targets. Aiming for these targets could create more reliable treatment avenues that interrupt key structural transitions in the virus’s lifecycle while also supporting the body’s natural immune response.
“A better understanding of the spike protein could complement current COVID-19 vaccines by informing new treatments and providing insights into potential drug design,” Bhowmik said.
Using the Nanoscale Molecular Dynamics, or NAMD, code on Summit, the nation’s most powerful supercomputer, the researchers simulated the spike proteins’ molecular structures for SARS-CoV-2 and three other human coronaviruses: SARS-CoV-1, MERS-CoV and HCoV-HKU1. After completing this unique and comprehensive comparison of four different spike proteins, they compared the components and behavior of SARS-CoV-2 with thousands of sample structures from the other viruses using a deep learning architecture called a convolutional variational autoencoder, or CVAE.
These efforts revealed previously unexplored regions of the coronavirus’s spike protein in which targeted medical intervention might prevent SARS-CoV-2 from infecting healthy cells. The researchers presented their findings at the IEEE Big Data Workshop for COVID-19, and their paper is published in the proceedings of the 2021 IEEE International Conference on Big Data.
Each spike protein contains three protein chains, or protomers, that are collectively known as the trimer. Each protomer comprises the amino-terminal domain, or NTD; the receptor binding domain, or RBD; and the S2 domain. The NTD and RBD are located in the S1 subunit of the spike protein, whereas the S2 domain resides in the S2 subunit.
“All of these coronaviruses have protomers that assemble to form a trimer, which means they have inherently flexible structures that can potentially be manipulated during assembly,” Chen said.
After confirming that SARS-CoV-2 has the same structural flexibility found in other coronaviruses, the team studied the spike protein in the protomer and the trimer state to pinpoint structural transitions that could be disrupted to destabilize the protein and negate its harmful effects.
The researchers discovered that two regions in the spike protein become vulnerable without the presence of certain stabilizing structures called beta sheets. These regions are the parts of the S2 domain that control the fusion of membranes between the virus and a host and the “hinge” that connects the S1 and S2 subunits.
Additionally, they found that antibodies recognized similar sites in the other coronavirus spike proteins, which led the team to conclude that interrupting the formation of beta sheets and preventing the protomers from interacting with one another could prevent the spike protein from forming a stable trimer and boost immune responses to SARS-CoV-2.
“We think these two regions are involved in helping the spike protein form the trimer,” Chen said. “Applying treatments to these regions could potentially prevent the virus from completing this process and infecting host cells.”
Many previous studies have focused exclusively on RBD because this domain binds directly to the angiotensin-converting enzyme 2, or ACE2, receptors in human cells. The scientists decided to expand their research to include other areas because, despite the crucial role it plays in infecting a host, RBD makes up a mere 15% of the spike protein.
“Studying the whole spike protein in detail allowed us to locate promising targets for medical approaches beyond those that have already been identified in RBD,” Bhowmik said.
The team was awarded a computing allocation on Summit, which is located at the Oak Ridge Leadership Computing Facility, through the COVID-19 High Performance Computing Consortium.
“At the beginning of this project, our back-of-the-envelope calculations revealed that we would need to run many simulations and generate an enormous amount of data to draw scientific conclusions,” Gounley said. “Summit provided the immense compute power we needed to handle that workload.”
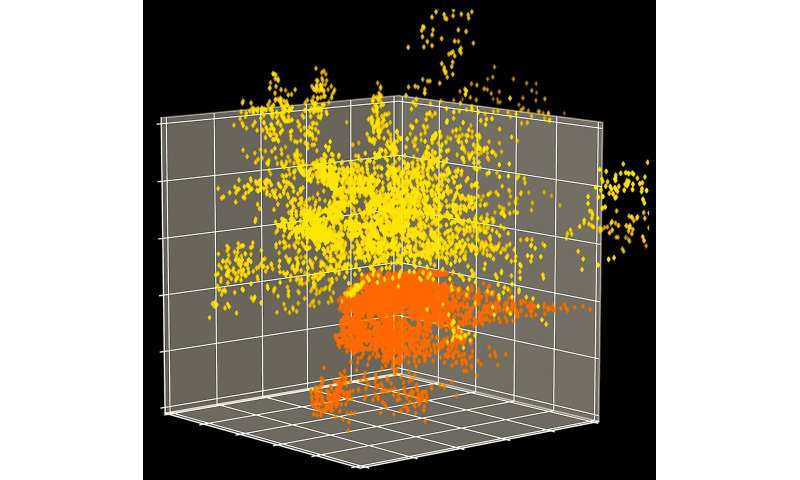
Although the researchers originally created their CVAE to characterize the structure of one small protein at a time, they developed a more advanced version to effectively examine multiple, much larger proteins simultaneously. Without this update, they could not have easily analyzed the simulation data because the SARS-CoV-2 spike protein is eight times larger than the proteins the CVAE was originally designed to study.
“This deep learning technique transforms massive amounts of data into manageable amounts of data while ensuring everything remains intact and accurate,” Bhowmik said. “We compressed the whole protein into a single dot that we can plot on a graph to see how the structure evolves over time.”
The team plans to continue studying the SARS-CoV-2 spike protein and anticipates that their methods could be applied to conduct additional spike protein analyses in other complex viruses.
This work was supported by DOE’s Office of Science, the Joint Design of Advanced Computing Solutions for Cancer program and the Exascale Computing Project. The researchers used resources of the OLCF, a DOE Office of Science user facility located at ORNL.
Source: Read Full Article
