Researchers at Cleveland Clinic have identified a new way that the SARS-CoV-2 virus hijacks its host’s translation machinery using elements within its genetic material and causes COVID-19, paving the way for future study.
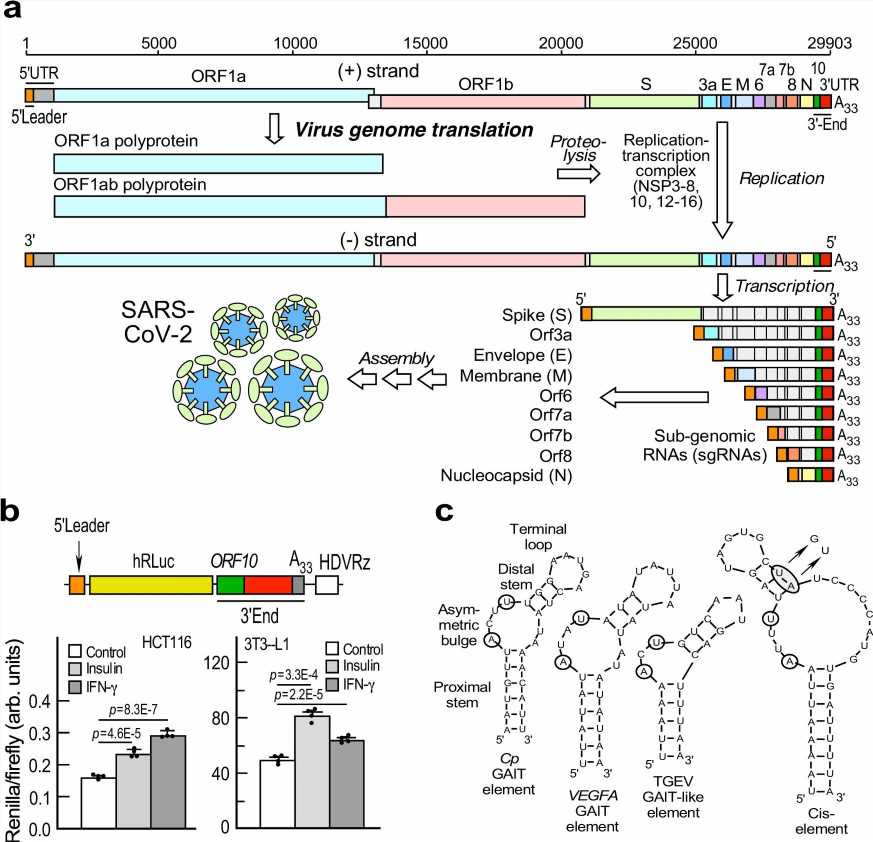
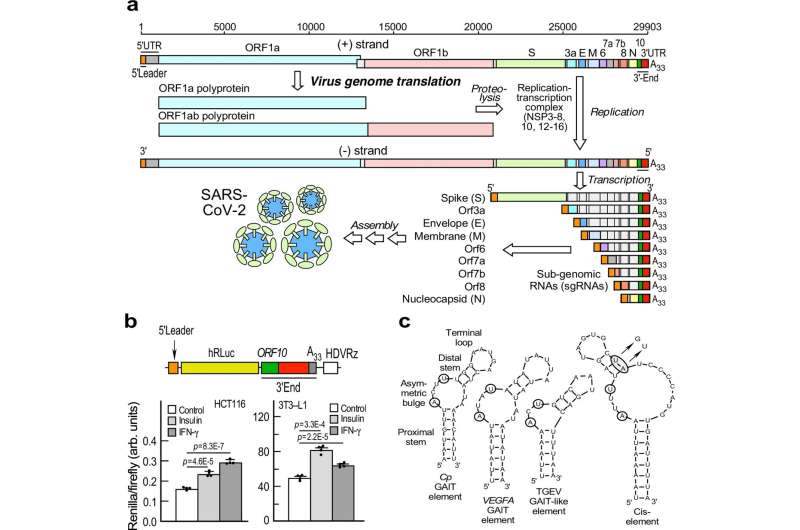
Viruses can’t replicate independently; they force infected host cells to read the genetic instructions in the virus’s RNA and produce proteins for the virus. A team led by Debjit Khan, Ph.D., from the laboratory of Paul Fox, Ph.D., report their newly discovered RNA element named SPEAR (sarbecoviral pan-end activating RNA) and a SPEAR-binding complex are crucial to how COVID-19 infects host cells to make this happen.
The team published their findings in a paper in Nature Communications, which also shares promising preclinical results from a new antiviral drug that blocks the SPEAR-binding complex.
“Current COVID-19 vaccines are a marvel of genetic engineering, but we must continue to learn as much as possible about the virus,” says Dr. Khan. “The current vaccines and treatments focus on the spike protein that binds to host cells, but that can mutate. This SPEAR element doesn’t change in the same way, so it’s important to investigate it to potentially add to our arsenal for fighting future outbreaks.”
When a new virus emerges, doctors notice and record trends among their patients—certain symptoms or groups of people who are affected more severely than others. Researchers can then go to work, using these observations to guide their projects.
SPEAR’s discovery also potentially brings us one step closer to why COVID-19 infections are more severe in patients with elevated insulin levels. Insulin, which helps the body regulate blood sugar, is dysregulated in diabetes and often elevated in patients with obesity. The study showed that insulin directly stimulates certain host proteins in fat cells to bind the SPEAR element and stimulate virus protein expression.
Using RNA to find answers about COVID-19
Investigating RNA is a critical component of understanding SARS-CoV-2 and developing treatments. The two investigators who completed the initial RNA research essential to developing the mRNA vaccine recently won the Nobel Prize in medicine.
Dr. Khan’s background is studying RNA in diseases like cancer and neurodevelopmental delay. As COVID-19 emerged, many RNA biologists in Dr. Fox’s lab began brainstorming how to investigate the virus’s RNA to learn more.
What started as a discussion led to collaboration with researchers in a lab led by Michaela Gack, Ph.D., Scientific Director of the Cleveland Clinic Florida Research & Innovation Center. Dr. Gack’s team in Infection Biology investigates the interactions between viruses and their hosts. GuanQun Liu, Ph.D., on Dr. Gack’s team completed some of the testing in facilities at the FRIC.
The team found that a section of the virus’s RNA folds into a distinct shape, mimicking a specific shape found in host RNA. That change allows the virus’s RNA to bind to host proteins and regulate which viral genes get made and at what levels. Four host enzymes were found to bind to the regulatory RNA and form the SPEAR-binding complex, which further enhanced virus protein synthesis.
“The proteins within the SPEAR complex we knew about, and we knew their functions within host cells. They’re essential for our health,” Dr. Khan says. “But we learned that the virus has figured out a way to use these for its own benefit.”
The study identified SPEAR and genes associated with the SPEAR-binding complex as well as other influences like insulin and circulating cytokine levels. Cytokines are immune regulators produced by the immune system which fight infection but also can cause inflammation. These discoveries have many implications outside of drug development, says Dr. Fox, Robert Canova Endowed Chair in Inflammation Research and a staff member in Cardiovascular & Metabolic Sciences of Lerner Research Institute.
More information:
Debjit Khan et al, A viral pan-end RNA element and host complex define a SARS-CoV-2 regulon, Nature Communications (2023). DOI: 10.1038/s41467-023-39091-3
Journal information:
Nature Communications
Source: Read Full Article