In a recent study published on the medRxiv* preprint server, researchers study the interaction between immunity to endemic coronaviruses (CoVs) and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Herein, the researchers used longitudinal epitope-resolved profiling of plasma antibodies before and after vaccination with the Moderna mRNA-1273 vaccine.
Study: COVID-19 vaccination recruits and matures cross-reactive antibodies to conserved epitopes in endemic coronavirus Spike proteins. Image Credit: createiveneko / Shutterstock.com
Background
Prior to the coronavirus disease 2019 (COVID-19) pandemic, humans already had immunity against four viruses of the coronaviridae family including HCoV-229E, HCoV-NL63, HCoV-OC43, and HCoV-HKU1. Human immunity is now increasing against SARS-CoV-2, a fifth virus of the coronaviridae family, through natural infection and worldwide vaccination efforts designed to immunize against its spike (S) protein.
Among COVID-19 vaccines, the Moderna mRNA-1273 is one of the most effective and widely- studied vaccines that generate robust T- and B- cell responses, including high titers of neutralizing antibodies against the S protein that correlates with protection against COVID-19. However, it remains poorly understood how pre-pandemic immunity to coronaviruses influences this newly acquired immunity against SARS-CoV-2.
About the study
In the present study, researchers designed and synthesized a peptide sequencing (PepSeq) library (HV2) that consisted of human virome-wide 15,000 30mer peptides representing 80 viruses that infect humans, including the four HCoVs and SARS-CoV/SARS-CoV-2.
The sequences were selected based on their antibody reactivity, including an empirical PepSeq screen conducted on a large peptide library, published epitopes of the immune epitope database, and homology with known reactive epitopes in related viral species.
Of the 421 peptides derived from five coronaviruses (CoV) species for the HV2 library, 174 originated from spike (S) sequences. Sixty other epitope sequences having two 30mer peptides containing SARS-CoV-2 S epitopes were subjected to saturation Alanine scanning.
PepSeq technology also allowed mapping of cross-reactive and non-cross-reactive S epitopes targeted by COVID-19 vaccination and for the researchers to study the evolution of corresponding antibody specificities over time.
The researchers recruited 21 subjects with no prior history of SARS-CoV-2 infection and undetectable anti-SARS-CoV-2 antibodies before receiving both doses of the mRNA-1273 vaccine. The blood samples were taken from each test subject at baseline and subsequently at approximately eight, 28 (preceding the second dose), and 140 days after complete vaccination.
All subjects showed a gradual increase in total S-binding and S-receptor-blocking antibodies, starting as early as eight days in some cases and uniformly reaching strong positivity by the final time point. Plasma samples were analyzed using the PepSeq platform, wherein the sample was incubated with a peptide library of thousands of DNA-barcoded peptides and immunoprecipitated onto protein G beads. Finally, the researchers identified the immunoglobulin G (IgG)-binding peptides through bulk amplification and high-throughput sequencing of the DNA tags.
Study findings
The cross-reactive epitopes of the S2 subunit, especially in the Fusion Peptide (FP) and Stem Helix (SH) region, are highly conserved and contain high sequence identity regions across SARS-CoV-2 and the four endemic HCoVs.
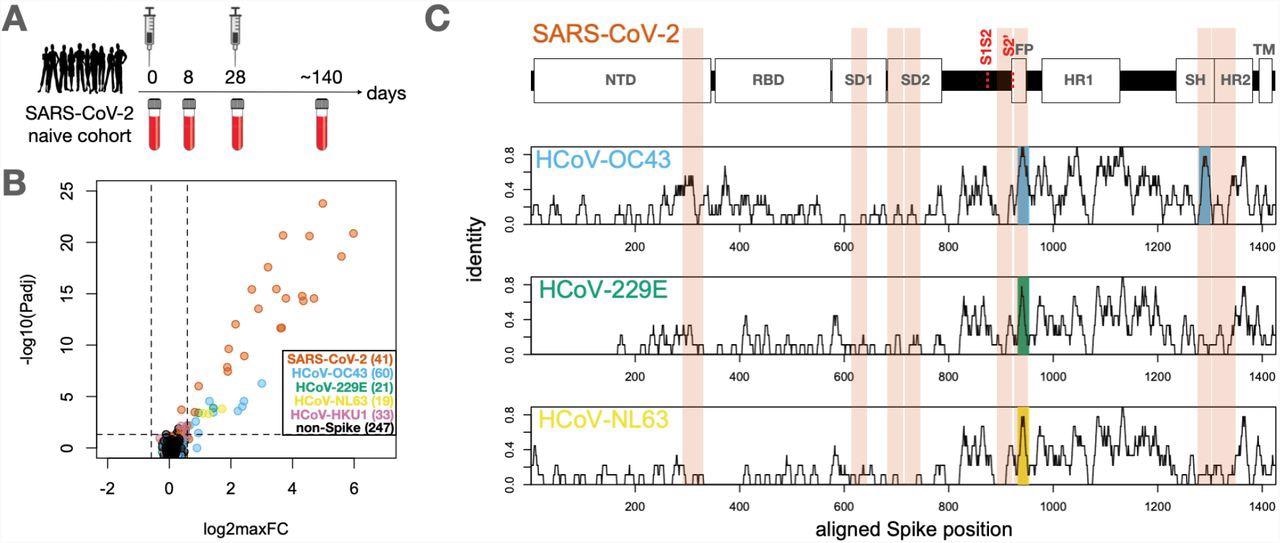
Identification of conserved and non-conserved antibody epitopes recognized following COVID-19 vaccination. A. A cohort of 21 subjects with no prior SARS-CoV-2 infection history and undetectable SARS-CoV-2 antibodies prior to vaccination received two doses of the mRNA-1273 vaccine and gave blood samples at days 0, ∼8, ∼28 and ∼140 days relative to the first dose. B. Plasma from each timepoint was analyzed by PepSeq with a 15,000-peptide human virome library (‘HV2’). Z-scores for the 421 HV2 peptides designed from members of the Coronaviridae family were analyzed across subjects and timepoints to identify peptides showing significant time-differential signals. Each dot represents an individual peptide: Y-axis shows the FDR-adjusted ANOVA P-value (Padj) across all timepoints, and X-axis shows maximum log2 fold change (log2maxFC), which was calculated by dividing maximum Z-score at day 8, 28, and 140 by the Z-score at day 0. Legend in the lower-right shows the number of peptides for each CoV Species. Dotted lines indicate thresholds at Padj=0.05 and maxFC=1.5. C. Mapping of differentially recognized peptides (passing both Padj and maxFC thresholds shown in B) to a multiple sequence alignment of Spike proteins from SARS-CoV-2, HCoV-OC43, HCoV-229E, and HCoV-NL63. Plots show structural features or cleavage sites (dotted red lines) of SARS-CoV-2 Spike (upper) and sliding-9mer window amino acid sequence identity to SARS-CoV-2 Spike for the three endemic Spike proteins (lower). The differentially-recognized peptides map to 8 epitopes in SARS-CoV-2 spike (vertical orange boxes), 2 epitopes in HCoV-OC43 (cyan), 1 epitope in HCoV-229E (gold), and 1 epitope in HCoV-NL63 (green). Epitopes detected in the three endemic CoV proteins all occur at two regions of high sequence conservation with SARS-CoV-2. The following features of Spike are highlighted: NTD=N-terminal Domain; RBD=Receptor Binding Domain; SD1=Subdomain 1; SD2=Subdomain 2; FP=Fusion Peptide; HR1=Heptad Repeat 1; SH=Stem Helix; HR2=Heptad Repeat 2; TM=Transmembrane region.
The serological results showed that COVID-19 vaccination elicited vigorous antibody responses against a range of S epitopes from SARS-CoV-2 and other HCoVs, which could be divided into two classes.
The first class of the antibody response was against minimally-conserved regions of the S protein that were undetectable until day 28 post-vaccination. This response strongly increased after the second dose. Notably, no antibody reactivity against homologous HCoV peptides for epitopes was observed under this class.
The second class of antibody responses was against the FP and SH of the highly-conserved S2 subunit of endemic HCoVs. The antibody reactivity to the SARS-CoV-2 sequences at these two sites followed the same kinetics as the first class, wherein they bound to the corresponding endemic homolog epitopes observed at day eight post-vaccination before decreasing between day 28 and day 140.
More importantly, antibody responses at these two epitope sites are dominated by cross-reactive antibodies. This pattern was observed at all post-vaccine timepoints for SH and the two earliest timepoints for FP.
The response to SARS-CoV-2 FP at day 140 was an exception, as it was substantially depleted by the SARS-CoV-2 sequence but not the endemic homolog. This observation indicates that a mono-specific reactivity was acquired at this epitope that reduced the endemic-specific binding of cross-reactive clones and led to the emergence of SARS-CoV-2-specific clones. Further, the core antibody recognition sites resided within high identity amino acid sequences in the betacoronavirus family in the case of SH, as observed in the mapping results.
The SARS-CoV-2 epitopes differed by two to three amino acids from their endemic homologs within each core recognition site, thereby indicating residues whose differential participation in antibody binding allowed the response to varying between species. The alanine-scanning results further supported this theory and revealed several instances where the antibody response showed selectively increased dependence on non-conserved residues while evolving from day 28 to day 140.
Conclusions
Overall, the results showed that, in addition to conferring protection against SARS-CoV-2, COVID-19 vaccination most likely boosts immunity against seasonal endemic CoVs through mediation by cross-reactive epitopes.
The protection against endemic HCoV infection when antibody-mediated was of limited duration; therefore, future epidemiological studies could determine its robustness. Nevertheless, it is likely that the cross-reactivity that is achieved through immunization or therapeutic strategies can elicit escape-resistant immunity across the CoV family.
Future studies tracking the sequences of individual cross-reactive antibody clones as their specificities evolve from endemic to SARS-CoV-2-dominant recognition, in combination with structural analysis, would also allow resolution of the molecular basis of the evolution between S2 homologs in detail.
Taken together, these observations suggest that antibodies with measurable neutralizing titers against multiple hCoVs may arise in humans following SARS-CoV-2 infection or vaccination. However, these responses have a limited role in boosting their overall immunity.
*Important notice
medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Elko, E. A., Nelson, G. A., Mead, H. L., et al. (2022). COVID-19 vaccination recruits and matures cross-reactive antibodies to conserved epitopes in endemic coronavirus Spike proteins. medRxiv. doi:10.1101/2022.01.24.22269542. https://www.medrxiv.org/content/10.1101/2022.01.24.22269542v1
Posted in: Medical Research News | Disease/Infection News | Pharmaceutical News
Tags: Alanine, Amino Acid, Antibodies, Antibody, Blood, Cell, Coronavirus, Coronavirus Disease COVID-19, DNA, Evolution, Helix, High-Throughput Sequencing, Homologous, immunity, Immunization, Immunoglobulin, Pandemic, Peptides, Protein, Receptor, Respiratory, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Syndrome, Vaccine, Virus

Written by
Neha Mathur
Neha Mathur has a Master’s degree in Biotechnology and extensive experience in digital marketing. She is passionate about reading and music. When she is not working, Neha likes to cook and travel.
Source: Read Full Article