Researchers tested a variety of RNA concentration and extraction methods and assays to develop a protocol for testing SARS-CoV-2 in wastewater, which may help improve COVID-19 surveillance.
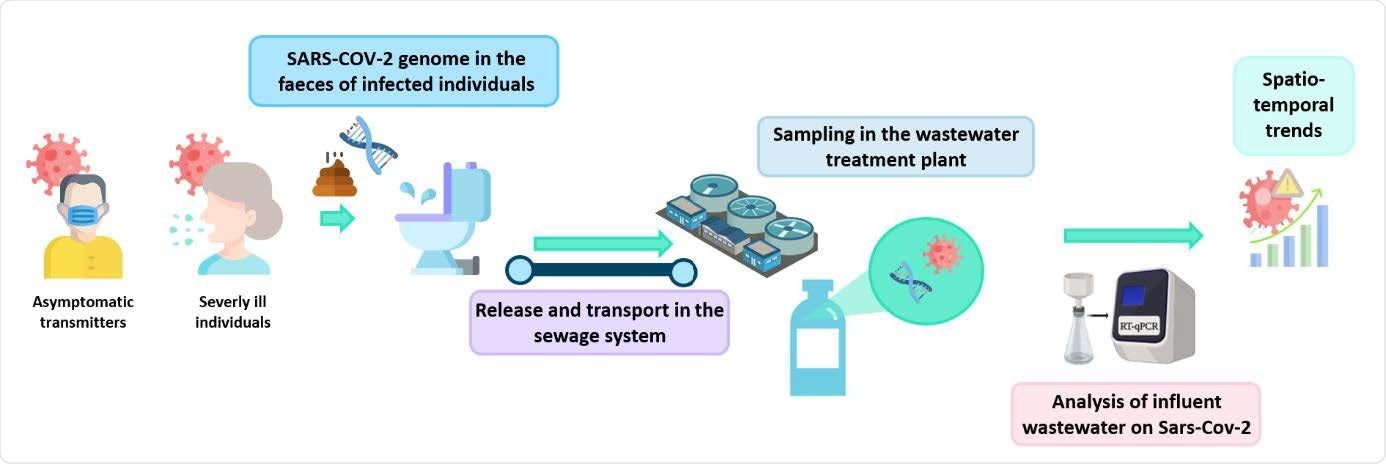
COVID-19, caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is usually monitored by diagnosing symptomatic patients and tracing their contacts. However, this may not be very accurate as it requires voluntary individual participation and does not include asymptomatic cases, making it challenging to control spread.
Testing wastewater is another method of monitoring the spread and circulation of infectious diseases. Disease markers such as viral genomes are released and transported in the wastewater. The abundance of pathogens in the wastewater can be used to infer the spread of disease.
Several studies have reported using wastewater to monitor the transmission of SARS-CoV-2. However, there are some challenges to doing this. Because wastewater has many other organic matter and metals, detection of low virus levels can be difficult. In addition, it is not known precisely in what form the SARS-CoV-2 virus is present in feces, such as virus particles, fragments, and so on, which may be broken down further during the transport of wastewater. So, it is essential to have reliable methods of concentration and analytical detection of viruses.
Testing strategies
In a research paper published on the medRxiv* preprint server, researchers from Belgium report a comparison of different bioanalytical methods for the concentration of SARS-CoV-2 RNA in wastewater.
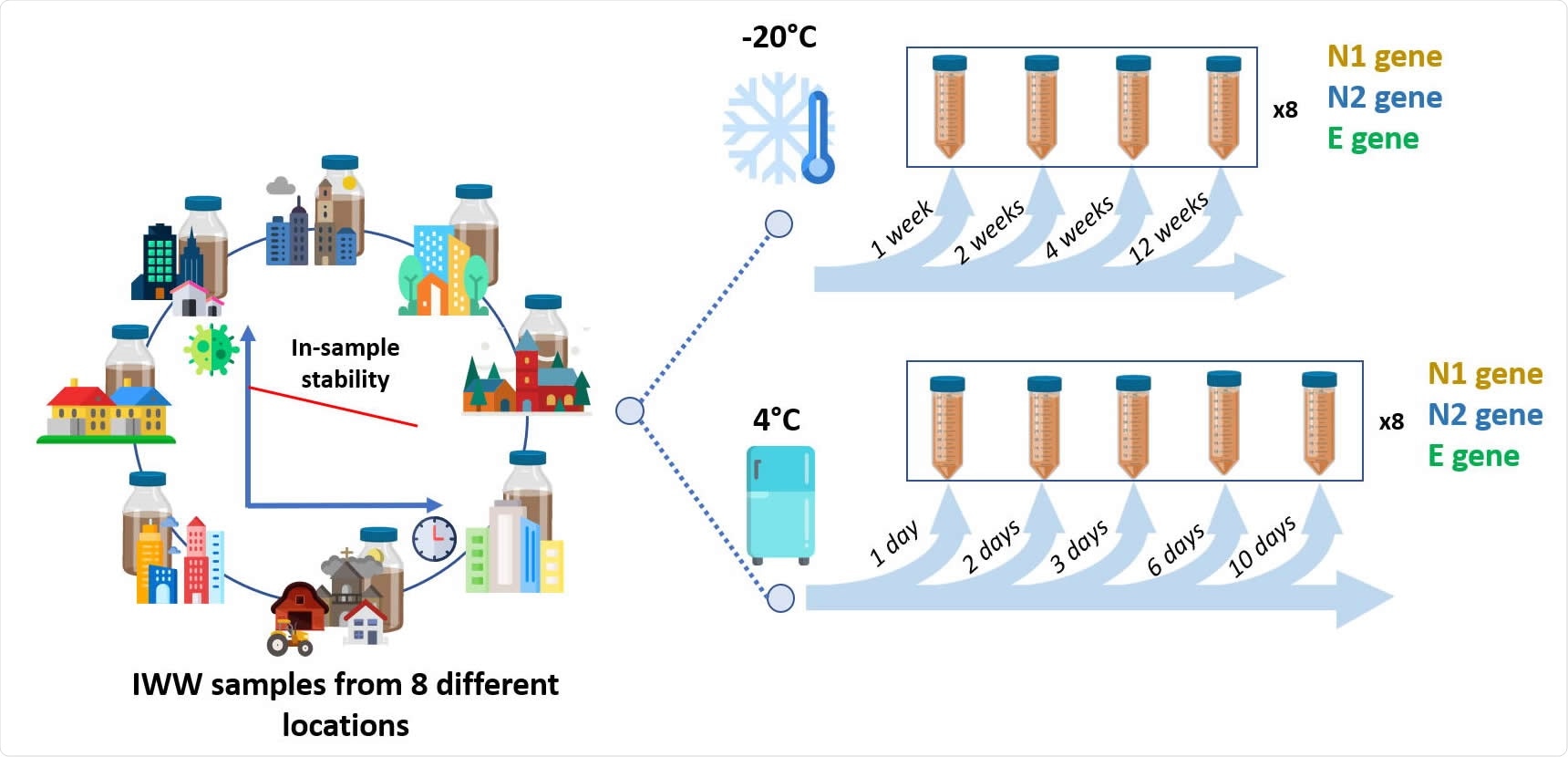
The team obtained wastewater samples from eight different wastewater treatment plants in Belgium and sanitary wastewater from a company that had a high number of COVID-19 cases. They used different ultrafiltration methods using several centrifugation devices and polyethylene glycol (PEG) precipitation to concentrate viral RNA. The method that gave the lowest and most reproducible levels of cycle threshold (Ct) for SARS-CoV-2 gene amplification was used for sample concentration.
The team also tested several commercial RNA extraction kits to choose the one that gave the lowest and most reproducible Ct values. They used both RT-PCR and digital PCR (dPCR) for the amplification of viral RNA. dPCR may be more useful for testing wastewater as it is less sensitive to PCR inhibitors present in wastewater and provides absolute quantification.
Improved analytical protocols
Based on the concentration results for samples collected in August 2020, when the number of cases was lower compared to those in the second wave, the team found PEG precipitation could not detect N1 and N2 genes and concentrations of porcine coronavirus were considerably lower. Hence, this method was not pursued further.
Upon testing the effect of different filter pore sizes and sample volumes, the team found that although smaller pore sizes can concentrate samples better, they often led to filter blockage and concentration of PCR inhibitors. So, they used a molecular weight cut-off of 50 to 100 kDa and sample volumes of 50 to 500 mL. Viral levels were too low to detect without concentration. Hence a concentration step was needed to detect SARS-CoV-2 in wastewater. In the final protocol, the researchers used ultracentrifugation with Centricon filters.
Although both the manual method of RNA extraction and the automated method using the Maxwell PureFood and GMO authentication kit were comparable for RNA extraction, the team chose the automated method for its high throughput.
The authors found both RT-PCR and dPCR gave the same concentration levels, indicating the sensitivity of both assays is similar. However, the advantage of dPCR is that it does not need a standard curve.
When water is transported in the sewer system, the SARS-CoV-2 genome may be broken into smaller RNA fragments. The team found N2 and E genes detected to be above the lower limit of quantification for more than 87.5% of the wastewater samples. However, there was increasing variability in the E gene with time. Since there was significant variability in the native concentration at the lower level of quantification, assessing stability is challenging.
Freezing samples decreased gene copies 10-fold, whereas detection levels were higher in samples kept at 4 °C. The authors suggest keeping samples at this temperature and analyzing them within three days of collection.
Despite developing better protocols, there is still significant variability in sample concentration methods as there is no external standard to compare with. Also, the state of the SARS-CoV-2 genome in wastewater is very uncertain because of potential degradation in the sewer system. Further research is needed to better understand these variabilities and the potential use of wastewater-based epidemiology for SARS-CoV-2 surveillance.
*Important Notice
medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Boogaerts, T. et al. (2021) An alternative approach for bioanalytical assay development for wastewater-based epidemiology of SARS-CoV-2. medRxiv. https://doi.org/10.1101/2021.02.12.21251626, https://www.medrxiv.org/content/10.1101/2021.02.12.21251626v1
Posted in: Device / Technology News | Medical Research News | Disease/Infection News
Tags: Coronavirus, Coronavirus Disease COVID-19, CT, Digital PCR, Epidemiology, Gene, Genes, Genome, High Throughput, Infectious Diseases, Research, Respiratory, RNA, RNA Extraction, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Syndrome, Virus

Written by
Lakshmi Supriya
Lakshmi Supriya got her BSc in Industrial Chemistry from IIT Kharagpur (India) and a Ph.D. in Polymer Science and Engineering from Virginia Tech (USA).
Source: Read Full Article
