Travel within your own country in response to the government announcing a national lockdown is directly linked to a sharp rise in COVID-19 infections, according to an international study led by Bangladesh and involving a researcher from the University of Bath.
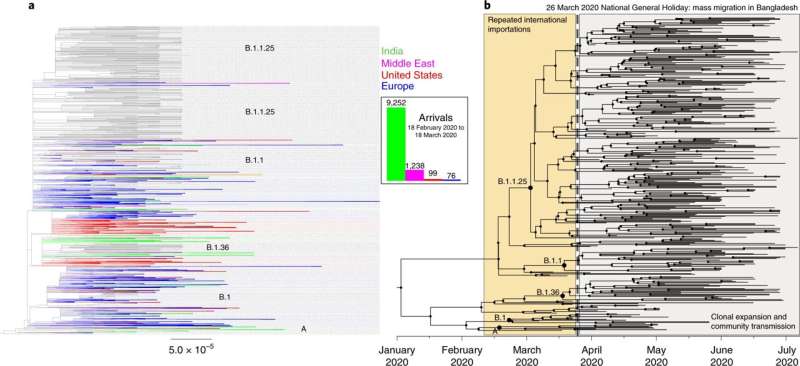
The study—the first of its kind to combine genome sequencing and mobile phone data—has found a direct link between mass migration from Dhaka, the capital of Bangladesh (an area of high infection), and the spread of coronavirus to the rest of the country after a stay-at-home order was announced during the nation’s first wave of infection in March 2020.
It is hoped that insights from this project will be used by other countries to track the virus and to inform the way governments respond when fresh outbreaks occur.
For the Bangladesh study, a consortium of scientists that included Dr. Lauren Cowley from the Department of Biology and Biochemistry at the University of Bath investigated the spatial spread of the coronavirus during Bangladesh’s first wave by analyzing the genome sequences of 391 viral samples collected from different parts of the country. This information was integrated with anonymised population-mobility data collected from Facebook and three mobile phone operators. This analysis gave the researchers insight into the evolution of the virus, and allowed them to track the different times and locations where new lineages of the virus appeared. It also enabled the authors to work directly with the government on the detection of variants of concern (Alpha and Beta) in Bangladesh.
The researchers established that much of the virus’ spread from the capital to all other areas of the country occurred during a three-day window between the government announcing a stay-at-home order and this order coming into effect (March 23–26, 2020). The population mobility data showed that a large proportion of Dhaka’s usual residents left the city for regional towns and villages in response to workplaces being closed.
As a result of the researchers’ findings, the government of Bangladesh restricted intercity travel during subsequent waves of infection, thereby making it harder for the virus to spread using the same channels as it did during the first wave.
Like Bangladesh, earlier in the pandemic the UK allowed time to pass between tighter COVID restrictions being announced and the new rules being imposed. On December 19 2020, in response to a localized spread of the new Alpha variant, the British prime minister declared that London and South East England would be entering a tier 4 “stay at home” alert level, however it wasn’t until December 21 that the restrictions came into effect. As a result of this time lag, many Londoners left the capital for other parts of the country, taking the virus with them.
Commenting, Dr. Cowley said: “We can apply lessons learnt from the Bangladesh study to the COVID situation in other countries. What we’ve learnt is that it’s best to shorten the time as much as possible between lockdown interventions being announced and imposed. Similar mass evacuations were seen in London in December 2020 to what we observed in Dhaka in late March 2020. These should be prevented as much as possible to limit the nationwide spread of the virus outside major capitals or other hotspots of infections.”
Professor Tahmina Shirin, director at IEDCR (Bangladesh’s national government public health department) and a senior author of the Bangladesh paper, said: “It is important that we continue to generate evidence to aid our policymakers to contain the spread of the virus successfully.
“Our consortium has provided many invaluable insights that helped the decision-makers to devise essential real-time policy decisions. These include the provision of mandatory quarantine and isolation of travelers arriving from countries where the variant of concern was dominating, imposing lockdown measures and restricting intercity movements of people, banning all international flights from high-risk countries to Bangladesh.”
Dr. Cowley, one of the first authors of the study, contributed to the project by training Bangladeshi scientists via Zoom in nanopore sequencing (a technology that enables DNA fragments to be analyzed) of the coronavirus. She also made a big contribution to the analysis of the sequences found in various parts of Bangladesh following the first wave of SARS-CoV-2 infection.
She said: “Of the samples sequenced during the first wave, we found that 85% fell into three dominant lineages. When examining our sequencing and mobility data in the context of the timeline of interventions that were put in place, we found that between March 23rd , when a national lockdown was announced, and March 26th, when the lockdown came into effect, there was a mass migration out of Dhaka to all areas of the country which resulted in the dispersal of these three lineages across Bangladesh.”
The first confirmed COVID-19 cases were recorded in Bangladesh on 8 March 2020. However, testing was slow to be implemented in regions outside of Dhaka, which made it hard to trace the spread of the virus during the first wave. In the week ending 11 April, new cases in Bangladesh grew by 1,155 percent, the highest increase in Asia at that time.
To date, Bangladesh has recorded nearly 1.5-million cases of coronavirus and almost 25,000 deaths during the pandemic.
The team of scientists involved in the study will continue to provide sequencing and analysis in Bangladesh. The researchers have been working closely with members of the Bangladesh government who are leading the public health response, to ensure policy decisions are guided by science and the latest genomic evidence in-country. This has led to a restriction on intercity travel being imposed in Bangladesh in response to the study’s findings from the first wave.
Thanks to their genomic surveillance work, the scientists were also able to reveal the dominance of the Beta variant during Bangladesh’s second wave of infection. Another positive outcome of this work is that the country has developed the capacity to respond to future outbreaks and epidemics.
Dr. Cowley said: “Several groups of Bangladeshi scientists have contributed to this and have collaboratively built a bank of Bangladesh SARS-CoV-2 sequences that is extremely important to public health officials tracking the outbreak there. Their genomic surveillance capacity, built during this pandemic, will be a tremendous asset to the country’s public health for many years to come. This is something that Bangladesh can be hugely proud of and that other countries wishing to implement genomic surveillance using comparatively low resources can learn from.”
Speaking of the emergence of new coronavirus variants and the associated risk, senior scientist at icddr,b (a Bangladesh-based international health research institute) and a senior author of the paper Dr. Firdausi Qadri said: “As more mutations accumulate in the SARS-CoV-2’s genetic code, there would be more variants of which some could have the strength to breakthrough natural or vaccine-induced immunity. This is already evident—Bangladesh has experienced the second wave with the Beta variant and is currently battling the third wave with the Delta variant. Real-time genomic surveillance is critical to understand the efficacy of these vaccines and to devise relevant strategies for Bangladesh and beyond.”
Commenting on the integration of genomic data with people’s movement data, Professor Caroline Buckee, study co-author and associate professor of epidemiology at the Harvard T.H. Chan School of Public Health, said: “These new data streams are valuable additions to traditional surveillance systems, and this study shows how they can be used during an emerging crisis to provide insights that are difficult to gain any other way. It also highlights the potential for interdisciplinary approaches like this to transform public health during this pandemic and in the future.”
This research is the result of a consortium comprised of the Wellcome Sanger Institute, the University of Bath, and Bangladesh-based Institutes that include the Institute of Epidemiology, Disease Control and Research (IEDCR), icddr,b (formerly International Centre for Diarrhoeal Disease Research) and the Institute for Developing Science and Health Initiatives (ideSHi), plus other local and international institutions.
Source: Read Full Article
