Researchers working across the USA, Ghana and South Africa have captured an instance of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) superinfection in an individual, simultaneously infected by two SARS-CoV-2 variants, Alpha and Epsilon during the second coronavirus disease 19 (COVID-19) wave in New York City in early 2021.
 Study: Capturing intrahost recombination of SARS-CoV-2 during superinfection with Alpha and Epsilon variants in New York City. Image Credit: Luca Lorenzelli/Shutterstock
Study: Capturing intrahost recombination of SARS-CoV-2 during superinfection with Alpha and Epsilon variants in New York City. Image Credit: Luca Lorenzelli/Shutterstock
Probing suggests that recombination, facilitated by superinfection, is perpetually occurring within SARS-CoV-2 infections, demonstrated by the occurrence of recombinant haplotypes at high frequency in the superinfected individual.
Background
A pre-print version of the research paper is available on the medRxiv* server while the article undergoes peer review.
Viral recombination occurs when at least two viral genomes, that are genetically distinct, co-infect or superinfect the same host cell and exchange genetic segments. It is a common evolutionary process in positive-strand RNA viruses, such as coronaviruses, to generate genetic diversity that allows them to overcome selective pressures while adapting to new environments and hosts.
Despite a regular occurrence during SARS-CoV-2 infections, recombination can be difficult to demonstrate in vivo unless it involves genetically distinct parental strains. Moreover, due to the short mean duration of SARS-CoV-2 infections, SARS-CoV-2 superinfections and recombinations have rarely been reported to date.
The team, in their recent study, characterize an instance of superinfection from January 2021, identified by the New York City (NYC) Department of Health and Mental Hygiene (DOHMH) and present evidence for recombination occurring within this superinfected individual, by providing an in vivo snapshot of this evolutionary process within SARS-CoV-2.
The study
The study was conducted on two SARS-CoV-2 positive nasopharyngeal swab samples, NYCPHL-002130 and NYCPHL-002461, that were submitted to NYC PHL (NYC Public Health Laboratory) for sequence analysis. In NYC, NYCPHL-002130 was the Index case belonging to the Alpha lineage of SARS-CoV-2 obtained from a sample drawn on 4 January 2021. Public health investigation on this index case revealed a history of recent travel to Ghana, where Alpha variant was already in circulation, and identified another contact case of an Alpha variant infection, sampled on 14 January 2021 (NYCPHL-002461).
Initial PCR screening by NYC PHL on index sample, NYCPHL-002130, demonstrated S gene target failure (SGTF) phenotype with the real-time PCR assay. However, sample collected from the contact partner (NYCPHL-002461) did not show the SGTF characteristic of the Alpha variant.
Later, genome sequencing on contact revealed substantial intrahost viral diversity- major and minor variants-within the viral genome, a possible signature of superinfection. To rule out any experimental or sequencing errors that could have led to this intrahost diversity, the sample NYCPHL-002461 was processed four times separately through independent reverse transcription, ARTIC PCR, library preparation, and sequencing reactions.
Genomic alignments to other publicly available alpha and Epsilon variant sequences were performed and separate maximum likelihood phylogenetic trees for both major and minor variants were inferred.
Molecular clock trees for the Alpha and Epsilon variants were inferred to determine whether the major and minor allelic variants were concomitant with the date of sampling.
To determine any onward transmission of major-minor strain recombinant, the team compared the sequences obtained in the current study with all SARS-CoV-2 genomes sequenced by the NYC PHL and Pandemic Response Lab (PRL) from the specimens collected from NYC residents.
Findings
Typical of the Alpha SARS-CoV-2 variant, NYCPHL-002130 exhibited S gene target failure (SGTF) phenotype in initial real-time screening. The viral genome from this index case showed limited intrahost viral diversity.
However, the contact partner NYCPHL-002461, did not exhibit the SGTF characteristic of the Alpha variant. And viral genome sequencing in this contact revealed substantial intrahost viral diversity, that indicated the instance of superinfection. A total of four replicate extractions, real-time PCR and sequencing runs demonstrated similar SGTF and confirmed the same signature of intrahost diversity.
The viral allelic frequencies (AF) in NYCPHL-002461 were segregated into four categories: shared, major strain, minor strain, and other. ‘Shared alleles’ referred to those present at 90% AF in three or more replicates (out of four replicates); ‘major strain alleles’ occurred at frequencies between 60 and 90% (≥3 replicates), with all diagnostic Alpha mutations in this set; ‘minor strain alleles’ that occurred at frequencies between 10 and 25% (≥3 replicates), with all except one diagnostic Epsilon mutation (A28272T mutation characteristic of Epsilon was absent in NYCPHL-002461) in this set. The ‘other’ category encompassed all other variable sites occurring at AF between 25% and 60% or those found in only one or two samples.
Root-to-tip regression analyses showed that the sampling date for NYCPHL-002461 was consistent with the molecular clock for both the major and minor strain sequences, and one would expect viruses of this degree of genetic divergence to have been circulating in mid-January 2021. The team also identified the genomes identical to the major variant circulating in both NYC (the NYCPHL-002130 index case) and in Ghana (EPI_ISL_944711) around the same time.
The minor variant belonging to the Epsilon variant lineage was found to be genetically distinct, with the closest Epsilon relatives sampled in California, the United Kingdom, and Cameroon. The most similar of these relatives was sampled in California that represented its direct ancestor on the phylogeny.
Cloning and sanger sequencing of longer genomic fragments provided greater resolution in the event of recombination. Later, to rule out the possibility of an in vitro recombination introduced by reverse-transcription and PCR amplification, which are part of both genome sequencing and cloning protocols, the recombinant haplotype frequencies across all the four extracts from NYCPHL-002461 were compared.
A consistent signal for recombination was observed in all the four whole genome sequencing analyses and in cloned-fragment analysis, all suggesting the same recombinant haplotypes present at high frequency.
We caution against assuming superinfection before potential issues of contamination, poor-quality sequencing, or bioinformatics errors have been appropriately dealt with”, advises the team.
No further transmission of the circulating recombinant was identified. The team points out that given that the initial index case and his contact were promptly given a COVID-19 diagnosis with advice to self-isolate by NYC DOHMH, the lack of onward transmission is not surprising.
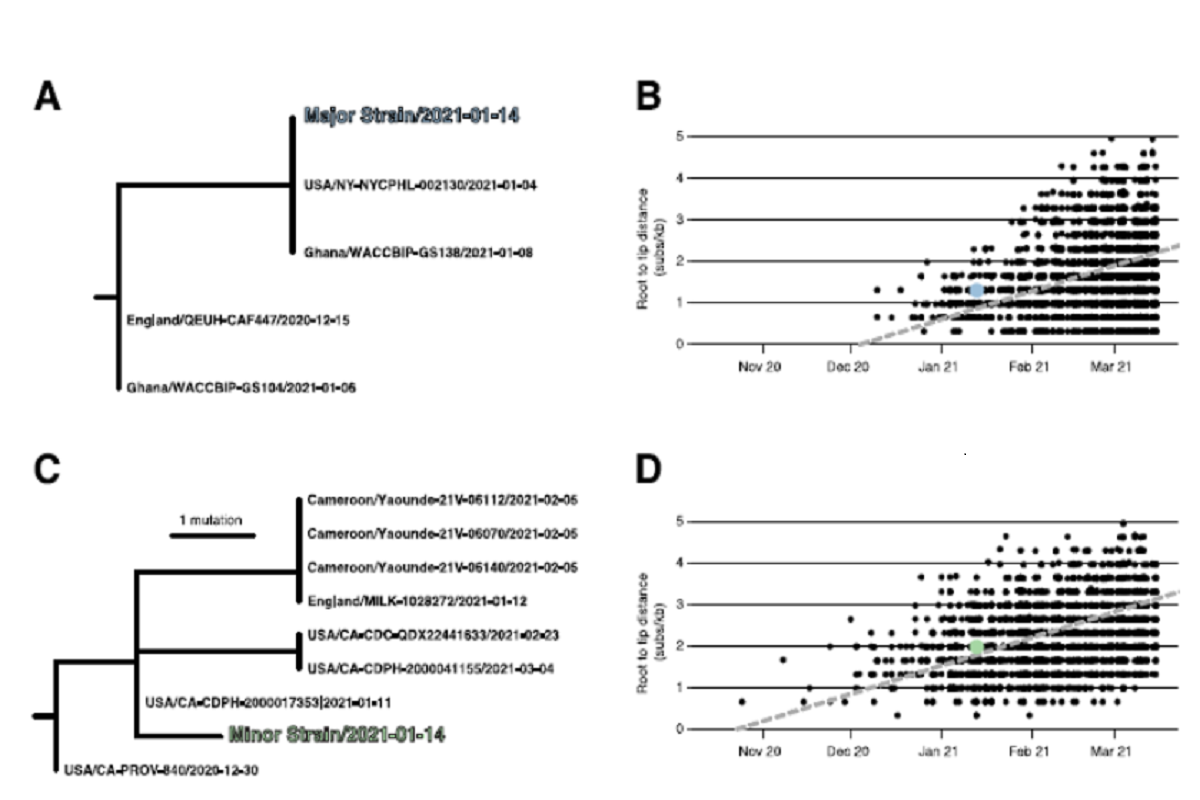 Figure 1: Phylogenetic consistency of major and minor 631 variants. (A) Phylogeny of B.1.1.7 immediate relatives, (B) Root-to-tip regression for B.1.1.7, (C) Phylogeny of B.1.429 immediate relatives, (D) Root-to-tip regression for B.1.429. NY-NYCPHL-0024661 is the genome deposited in GISAID from the case of putative superinfection.
Figure 1: Phylogenetic consistency of major and minor 631 variants. (A) Phylogeny of B.1.1.7 immediate relatives, (B) Root-to-tip regression for B.1.1.7, (C) Phylogeny of B.1.429 immediate relatives, (D) Root-to-tip regression for B.1.429. NY-NYCPHL-0024661 is the genome deposited in GISAID from the case of putative superinfection.
Because recombinant viruses can be successfully generated and transmitted between humans, this finding underscores their potential relevance to the future of the COVID-19 pandemic”, highlights the team.
Conclusion
The occurrence of high frequency genetically variable recombinant haplotypes within a single superinfected individual suggests that recombination is constantly occurring within SARS-CoV-2 infections.
Reduced incidence due to vaccine-induced and naturally-acquired immunity would lower the opportunity for superinfection, and the homogenizing effect of variant-driven selective sweeps (as seen in the Delta and Omicron variants) will lessen the potential for biological innovation in a recombinant genome.”
However, molecular surveillance for the emergence of a recombinant SARS-CoV-2 variant should actively continue to timely inform the public health authorities.
*Important notice
medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Wertheim J., O., Wang J., C., & Leelawong M., et al. (2022) Capturing intrahost recombination of SARS-CoV-2 during superinfection with Alpha and Epsilon variants in New York City. medRxiv. doi: https://doi.org/10.1101/2022.01.18.22269300, https://www.medrxiv.org/content/10.1101/2022.01.18.22269300v1
Posted in: Medical Science News | Medical Research News | Disease/Infection News
Tags: Assay, Bioinformatics, Cell, Cloning, Contamination, Coronavirus, Coronavirus Disease COVID-19, covid-19, Diagnostic, Frequency, Gene, Genetic, Genome, Genomic, Hygiene, immunity, in vivo, Laboratory, Library Preparation, Mutation, Nasopharyngeal, Omicron, Pandemic, Phenotype, Phylogeny, Public Health, Research, Respiratory, RNA, Sanger sequencing, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Syndrome, Transcription, Vaccine, Whole Genome Sequencing

Written by
Namita Mitra
After earning a bachelor’s degree in Veterinary Sciences and Animal Health (BVSc) in 2013, Namita went on to pursue a Master of Veterinary Microbiology from GADVASU, India. Her Master’s research on the molecular and histopathological diagnosis of avian oncogenic viruses in poultry brought her two national awards. In 2013, she was conferred a doctoral degree in Animal Biotechnology that concluded with her research findings on expression profiling of apoptosis-associated genes in canine mammary tumors. Right after her graduation, Namita worked as Assistant Professor of Animal Biotechnology and taught the courses of Animal Cell Culture, Animal Genetic Engineering, and Molecular Immunology.
Source: Read Full Article
